(This article was first published on R snippets, and kindly contributed to R-bloggers)
Recently I had several discussions about using for loops in GNU R and how they compare to *apply family in terms of speed. I have not seen a direct benchmark comparing them so I decided to execute one (warning: some of the code presented today takes long time to execute).First I have started by comparing the speed of assignment operator for lists vs. numeric vectors in standard for loops. Here is the code:
speed.test <- function(n) {The picture showing the result of the comparison is the following:
gc()
x1 <- numeric(n)
x2 <- vector(n, mode = "list")
c(system.time(for (i in 1:n) { x1[i] <- i })[3],
system.time(for (i in 1:n) { x2[[i]] <- i })[3])
}
n <- seq(10 ^ 4, 10 ^ 6, len = 5)
result <- t(sapply(n, speed.test))
par(mar=c(4.5, 4.5, 1, 1))
matplot(n / 1000, result, type = "l" , col = 1:2, lty = 1,
xlab = "n ('000)", ylab = "time")
legend("topleft", legend = c("numeric", "list"),
col = 1:2, lty = 1)
As we can see - operation numeric vectors are significantly faster than list, especially for large vector sizes.
But how does this relate to *apply family of functions? The issue is that the workhorse function there is lapply and it works on lists. Other functions from this family call lapply internally.
So I have run the second test comparing: (a) lapply, (b) for loop working on lists and (c) for loop working on numeric vectors. Here is the code:
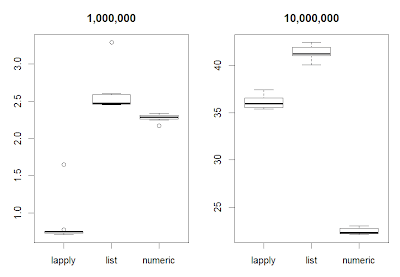
aworker <- function(n) {On the picture below we can see the result. For 1,000,000 elements in a vector lapply is the fastest. The reason it that it executes looping in compiled C code. However for 10,000,000 elements for loop using numeric vector is faster as it avoids conversion to list.
r <- lapply(1:n, identity)
return(NULL)
}
lworker <- function(n) {
r <- vector(n, mode = "list")
for (i in 1:n) {
r[[i]] <- identity(i)
}
return(NULL)
}
nworker <- function(n) {
r <- numeric(n)
for (i in 1:n) {
r[i] <- identity(i)
}
return(NULL)
}
run <- function(n, worker) {
gc()
unname(system.time(worker(n))[3])
}
compare <- function(n) {
c(lapply = run(n, aworker),
list = run(n, lworker),
numeric = run(n, nworker))
}
n <- rep(c(10 ^ 6, 10 ^ 7), 10)
result <- t(sapply(n, compare))
par(mfrow = c(1,2), mar = c(3,3,3,1))
for (i in n[1:2]) {
boxplot(result[n == i,],
main = format(i, scientific = F, big.mark = ","))
}
Of course probably on other machines than my notebook the difference in speed would manifest itself for other number of elements in a vector.
However one can draw a general conclusion: if you have large AND numeric vectors and need to do a lot of number crunching for loop will be faster than lapply.
To leave a comment for the author, please follow the link and comment on his blog: R snippets.
R-bloggers.com offers daily e-mail updates about R news and tutorials on topics such as: visualization (ggplot2, Boxplots, maps, animation), programming (RStudio, Sweave, LaTeX, SQL, Eclipse, git, hadoop, Web Scraping) statistics (regression, PCA, time series,ecdf, trading) and more...